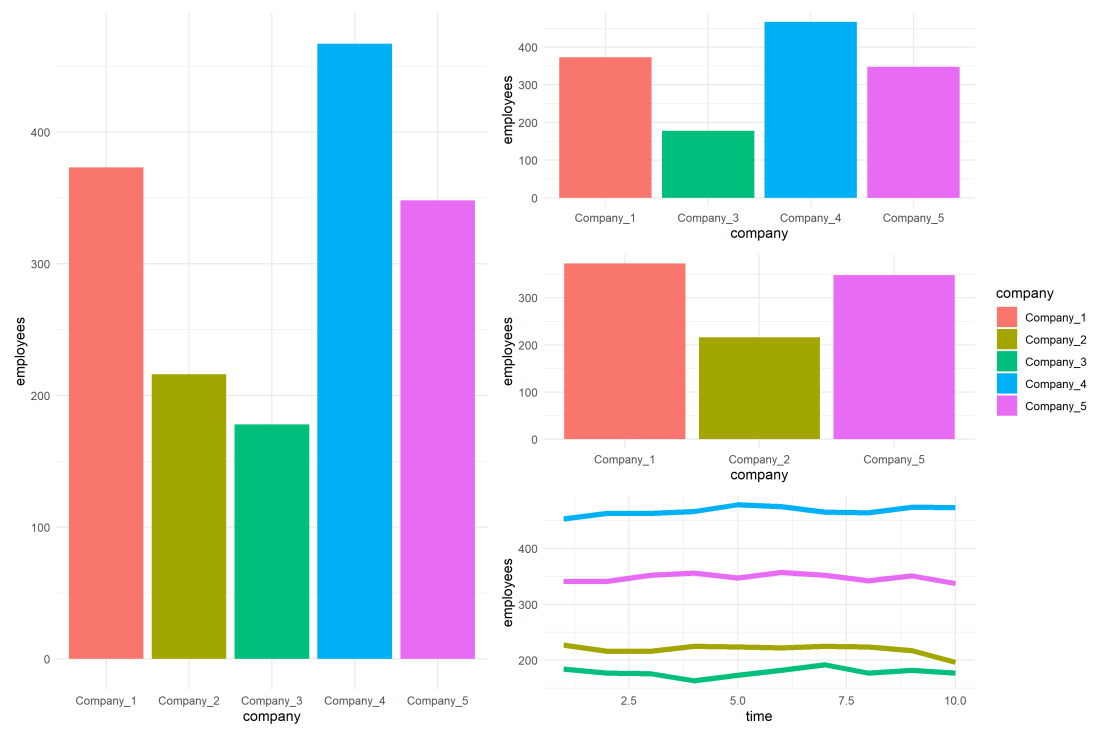
One best practice in visualization is to make your color scheme consistent across figures.
For instance, if you’re making multiple plots of the dataset — say a group of 5 companies — you want to have each company have the same, consistent coloring across all these plots.
R has some great data visualization capabilities. Particularly the ggplot2 package makes it so easy to spin up a good-looking visualization quickly.
The default in R is to look at the number of groups in your data, and pick “evenly spaced” colors across a hue color wheel. This looks great straight out of the box:
# install.packages('ggplot2')
library(ggplot2)
theme_set(new = theme_minimal()) # sets a default theme
set.seed(1) # ensure reproducibility
# generate some data
n_companies = 5
df1 = data.frame(
company = paste('Company', seq_len(n_companies), sep = '_'),
employees = sample(50:500, n_companies),
stringsAsFactors = FALSE
)
# make a simple column/bar plot
ggplot(data = df1) +
geom_col(aes(x = company, y = employees, fill = company))
However, it can be challenging is to make coloring consistent across plots.
For instance, suppose we want to visualize a subset of these data points.
index_subset1 = c(1, 3, 4, 5) # specify a subset
# make a plot using the subsetted dataframe
ggplot(data = df1[index_subset1, ]) +
geom_col(aes(x = company, y = employees, fill = company))
As you can see the color scheme has now changed. With one less group / company, R now picks 4 new colors evenly spaced around the color wheel. All but the first are different to the original colors we had for the companies.
One way to deal with this in R and ggplot2, is to add a scale_* layer to the plot.
Here we manually set Hex color values in the scale_fill_manual function. These hex values I provided I know to be the default R values for four groups.
# install.packages('scales')
# the hue_pal function from the scales package looks up a number of evenly spaced colors
# which we can save as a vector of character hex values
default_palette = scales::hue_pal()(5)
# these colors we can then use in a scale_* function to manually override the color schema
ggplot(data = df1[index_subset1, ]) +
geom_col(aes(x = company, y = employees, fill = company)) +
scale_fill_manual(values = default_palette[-2]) # we remove the element that belonged to company 2
As you can see, the colors are now aligned with the previous schema. Only Company 2 is dropped, but all other companies retained their color.

However, this was very much hard-coded into our program. We had to specify which company to drop using the default_palette[-2].
If the subset changes, which often happens in real life, our solution will break as the values in the palette no longer align with the groups R encounters:
index_subset2 = c(1, 2, 5) # but the subset might change
# and all manually-set colors will immediately misalign
ggplot(data = df1[index_subset2, ]) +
geom_col(aes(x = company, y = employees, fill = company)) +
scale_fill_manual(values = default_palette[-2])
Fortunately, R is a smart language, and you can work your way around this!
All we need to do is created, what I call, a named-color palette!
It’s as simple as specifying a vector of hex color values! Alternatively, you can use the grDevices::rainbow or grDevices::colors() functions, or one of the many functions included in the scales package
# you can hard-code a palette using color strings
c('red', 'blue', 'green')
# or you can use the rainbow or colors functions of the grDevices package
rainbow(n_companies)
colors()[seq_len(n_companies)]
# or you can use the scales::hue_pal() function
palette1 = scales::hue_pal()(n_companies)
print(palette1)[1] "#F8766D" "#A3A500" "#00BF7D" "#00B0F6" "#E76BF3"
Now we need to assign names to this vector of hex color values. And these names have to correspond to the labels of the groups that we want to colorize.
You can use the names function for this.
names(palette1) = df1$company
print(palette1)Company_1 Company_2 Company_3 Company_4 Company_5 "#F8766D" "#A3A500" "#00BF7D" "#00B0F6" "#E76BF3"
But I prefer to use the setNames function so I can do the inititialization, assignment, and naming simulatenously. It’s all the same though.
palette1_named = setNames(object = scales::hue_pal()(n_companies), nm = df1$company)
print(palette1_named)Company_1 Company_2 Company_3 Company_4 Company_5 "#F8766D" "#A3A500" "#00BF7D" "#00B0F6" "#E76BF3"
With this named color vector and the scale_*_manual functions we can now manually override the fill and color schemes in a flexible way. This results in the same plot we had without using the scale_*_manual function:
ggplot(data = df1) +
geom_col(aes(x = company, y = employees, fill = company)) +
scale_fill_manual(values = palette1_named)
However, now it does not matter if the dataframe is subsetted, as we specifically tell R which colors to use for which group labels by means of the named color palette:
# the colors remain the same if some groups are not found
ggplot(data = df1[index_subset1, ]) +
geom_col(aes(x = company, y = employees, fill = company)) +
scale_fill_manual(values = palette1_named)
# and also if other groups are not found
ggplot(data = df1[index_subset2, ]) +
geom_col(aes(x = company, y = employees, fill = company)) +
scale_fill_manual(values = palette1_named)
Once you are aware of these superpowers, you can do so much more with them!
How about highlighting a specific group?
Just set all the other colors to ‘grey’…
# lets create an all grey color palette vector
palette2 = rep('grey', times = n_companies)
palette2_named = setNames(object = palette2, nm = df1$company)
print(palette2_named)Company_1 Company_2 Company_3 Company_4 Company_5 "grey" "grey" "grey" "grey" "grey"
# this looks terrible in a plot
ggplot(data = df1) +
geom_col(aes(x = company, y = employees, fill = company)) +
scale_fill_manual(values = palette2_named)
… and assign one of the company’s colors to be a different color
# override one of the 'grey' elements using an index by name
palette2_named['Company_2'] = 'red'
print(palette2_named)Company_1 Company_2 Company_3 Company_4 Company_5 "grey" "red" "grey" "grey" "grey"
# and our plot is professionally highlighting a certain group
ggplot(data = df1) +
geom_col(aes(x = company, y = employees, fill = company)) +
scale_fill_manual(values = palette2_named)
We can apply these principles to other types of data and plots.
For instance, let’s generate some time series data…
timepoints = 10
df2 = data.frame(
company = rep(df1$company, each = timepoints),
employees = rep(df1$employees, each = timepoints) + round(rnorm(n = nrow(df1) * timepoints, mean = 0, sd = 10)),
time = rep(seq_len(timepoints), times = n_companies),
stringsAsFactors = FALSE
)… and visualize these using a line plot, adding the color palette in the same way as before:
ggplot(data = df2) +
geom_line(aes(x = time, y = employees, col = company), size = 2) +
scale_color_manual(values = palette1_named)
If we miss one of the companies — let’s skip Company 2 — the palette makes sure the others remained colored as specified:
ggplot(data = df2[df2$company %in% df1$company[index_subset1], ]) +
geom_line(aes(x = time, y = employees, col = company), size = 2) +
scale_color_manual(values = palette1_named)
Also the highlighted color palete we used before will still work like a charm!
ggplot(data = df2) +
geom_line(aes(x = time, y = employees, col = company), size = 2) +
scale_color_manual(values = palette2_named)
Now, let’s scale up the problem! Pretend we have not 5, but 20 companies.
The code will work all the same!
set.seed(1) # ensure reproducibility
# generate new data for more companies
n_companies = 20
df1 = data.frame(
company = paste('Company', seq_len(n_companies), sep = '_'),
employees = sample(50:500, n_companies),
stringsAsFactors = FALSE
)
# lets create an all grey color palette vector
palette2 = rep('grey', times = n_companies)
palette2_named = setNames(object = palette2, nm = df1$company)
# highlight one company in a different color
palette2_named['Company_2'] = 'red'
print(palette2_named)
# make a bar plot
ggplot(data = df1) +
geom_col(aes(x = company, y = employees, fill = company)) +
scale_fill_manual(values = palette2_named) +
theme(axis.text.x = element_text(angle = 45, hjust = 1, vjust = 1)) # rotate and align the x labels
Also for the time series line plot:
timepoints = 10
df2 = data.frame(
company = rep(df1$company, each = timepoints),
employees = rep(df1$employees, each = timepoints) + round(rnorm(n = nrow(df1) * timepoints, mean = 0, sd = 10)),
time = rep(seq_len(timepoints), times = n_companies),
stringsAsFactors = FALSE
)
ggplot(data = df2) +
geom_line(aes(x = time, y = employees, col = company), size = 2) +
scale_color_manual(values = palette2_named)
The possibilities are endless; the power is now yours!
Just think at the efficiency gain if you would make a custom color palette, with for instance your company’s brand colors!
For more R tricks to up your programming productivity and effectiveness, visit the R tips and tricks page!