Laura DeCicco found that non-R users keep asking her what her box plots exactly mean or demonstrate. In a recent blog post, she therefore breaks down the calculations into easy-to-follow chunks of code. Even better, she included the source code to make boxplots that come with a very elaborate default legend:

As you can see, the above contains much more and easier to understand information than the original ggplot2 boxplot below.

Laura wrote the custom function ggplot_box_legend() (see source code below and in Laura’s blog), which uses the cowplot package to paste the explanation to the box plot. All you need to do is call the legend function just before you run your ggplot2 boxplot call.
ggplot_box_legend <- function(family = "serif"){
# Create data to use in the boxplot legend:
set.seed(100)
sample_df <- data.frame(parameter = "test",
values = sample(500))
# Extend the top whisker a bit:
sample_df$values[1:100] <- 701:800
# Make sure there's only 1 lower outlier:
sample_df$values[1] <- -350
# Function to calculate important values:
ggplot2_boxplot <- function(x){
quartiles <- as.numeric(quantile(x,
probs = c(0.25, 0.5, 0.75)))
names(quartiles) <- c("25th percentile",
"50th percentile\n(median)",
"75th percentile")
IQR <- diff(quartiles[c(1,3)])
upper_whisker <- max(x[x < (quartiles[3] + 1.5 * IQR)])
lower_whisker <- min(x[x > (quartiles[1] - 1.5 * IQR)])
upper_dots <- x[x > (quartiles[3] + 1.5*IQR)]
lower_dots <- x[x < (quartiles[1] - 1.5*IQR)]
return(list("quartiles" = quartiles,
"25th percentile" = as.numeric(quartiles[1]),
"50th percentile\n(median)" = as.numeric(quartiles[2]),
"75th percentile" = as.numeric(quartiles[3]),
"IQR" = IQR,
"upper_whisker" = upper_whisker,
"lower_whisker" = lower_whisker,
"upper_dots" = upper_dots,
"lower_dots" = lower_dots))
}
# Get those values:
ggplot_output <- ggplot2_boxplot(sample_df$values)
# Lots of text in the legend, make it smaller and consistent font:
update_geom_defaults("text",
list(size = 3,
hjust = 0,
family = family))
# Labels don't inherit text:
update_geom_defaults("label",
list(size = 3,
hjust = 0,
family = family))
# Create the legend:
# The main elements of the plot (the boxplot, error bars, and count)
# are the easy part.
# The text describing each of those takes a lot of fiddling to
# get the location and style just right:
explain_plot <- ggplot() + stat_boxplot(data = sample_df, aes(x = parameter, y=values), geom ='errorbar', width = 0.3) + geom_boxplot(data = sample_df, aes(x = parameter, y=values), width = 0.3, fill = "lightgrey") + geom_text(aes(x = 1, y = 950, label = "500"), hjust = 0.5) + geom_text(aes(x = 1.17, y = 950, label = "Number of values"), fontface = "bold", vjust = 0.4) + theme_minimal(base_size = 5, base_family = family) + geom_segment(aes(x = 2.3, xend = 2.3, y = ggplot_output[["25th percentile"]], yend = ggplot_output[["75th percentile"]])) + geom_segment(aes(x = 1.2, xend = 2.3, y = ggplot_output[["25th percentile"]], yend = ggplot_output[["25th percentile"]])) + geom_segment(aes(x = 1.2, xend = 2.3, y = ggplot_output[["75th percentile"]], yend = ggplot_output[["75th percentile"]])) + geom_text(aes(x = 2.4, y = ggplot_output[["50th percentile\n(median)"]]), label = "Interquartile\nrange", fontface = "bold", vjust = 0.4) + geom_text(aes(x = c(1.17,1.17), y = c(ggplot_output[["upper_whisker"]], ggplot_output[["lower_whisker"]]), label = c("Largest value within 1.5 times\ninterquartile range above\n75th percentile", "Smallest value within 1.5 times\ninterquartile range below\n25th percentile")), fontface = "bold", vjust = 0.9) + geom_text(aes(x = c(1.17), y = ggplot_output[["lower_dots"]], label = "Outside value"), vjust = 0.5, fontface = "bold") + geom_text(aes(x = c(1.9), y = ggplot_output[["lower_dots"]], label = "-Value is >1.5 times and"),
vjust = 0.5) +
geom_text(aes(x = 1.17,
y = ggplot_output[["lower_dots"]],
label = "<3 times the interquartile range\nbeyond either end of the box"),
vjust = 1.5) +
geom_label(aes(x = 1.17, y = ggplot_output[["quartiles"]],
label = names(ggplot_output[["quartiles"]])),
vjust = c(0.4,0.85,0.4),
fill = "white", label.size = 0) +
ylab("") + xlab("") +
theme(axis.text = element_blank(),
axis.ticks = element_blank(),
panel.grid = element_blank(),
aspect.ratio = 4/3,
plot.title = element_text(hjust = 0.5, size = 10)) +
coord_cartesian(xlim = c(1.4,3.1), ylim = c(-600, 900)) +
labs(title = "EXPLANATION")
return(explain_plot)
}
ggplot_box_legend()


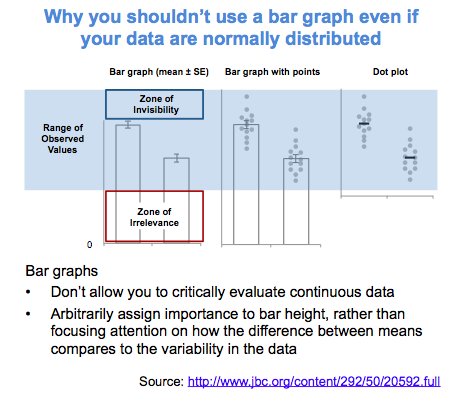
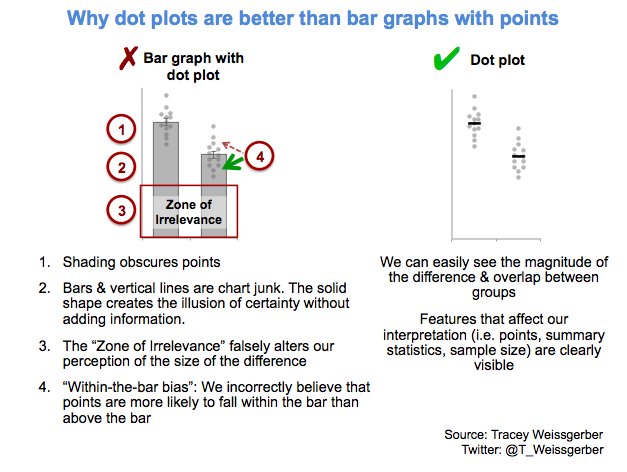
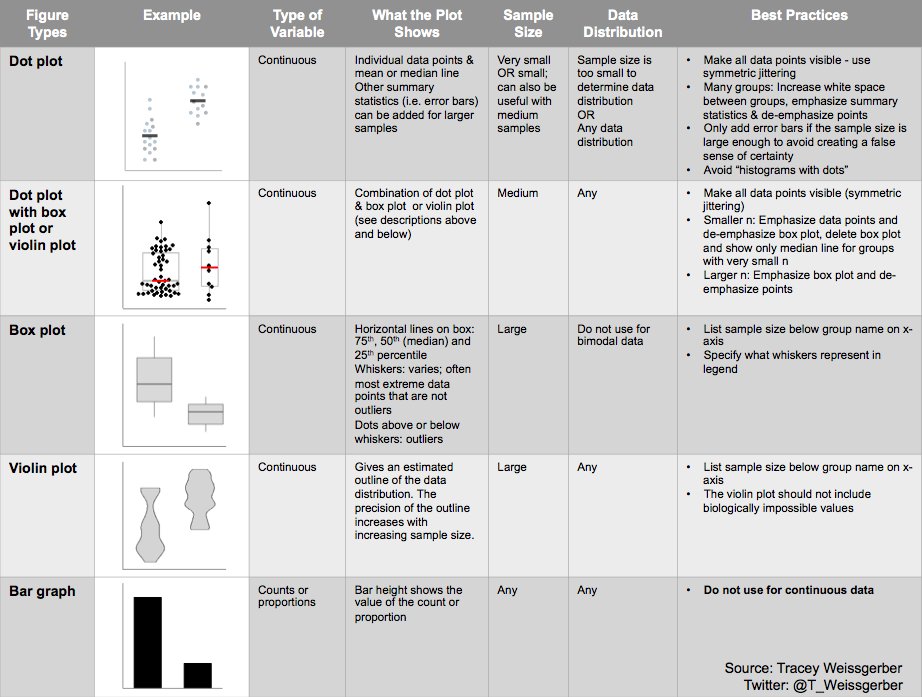
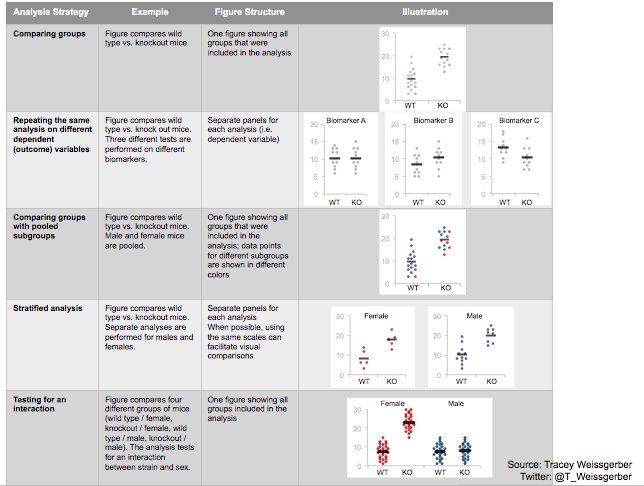



 Alternatively, scatterplots can be easily generated. Displaying missings at 10 percent below the minimum of the airquality dataset. Scatterplots of ozone and solar radiation (A), and ozone and temperature (B). These plots demonstrate that there are missings in ozone and solar radiation, but not in temperature.
Alternatively, scatterplots can be easily generated. Displaying missings at 10 percent below the minimum of the airquality dataset. Scatterplots of ozone and solar radiation (A), and ozone and temperature (B). These plots demonstrate that there are missings in ozone and solar radiation, but not in temperature.




