I recently came across this lovely article where Ali Spittel provides 7 tips for writing cleaner JavaScript code. Enthusiastic about her guidelines, I wanted to translate them to the R programming environment. However, since R is not an object-oriented programming language, not all tips were equally relevant in my opinion. Here’s what really stood out for me.

1. Use clear variable and function names
Suppose we want to create our own custom function to derive the average value of a vector v (please note that there is a base::mean function to do this much more efficiently). We could use the R code below to compute that the average of vector 1 through 10 is 5.5.
avg <- function(v){
s = 0
for(i in seq_along(v)) {
s = s + v[i]
}
return(s / length(v))
}
avg(1:10) # 5.5
However, Ali rightfully argues that this code can be improved by making the variable and function names much more explicit. For instance, the refigured code below makes much more sense on a first look, while doing exactly the same.
averageVector <- function(vector){
sum = 0
for(i in seq_along(vector)){
sum = sum + vector[i]
}
return(sum / length(vector))
}
averageVector(1:10) #5.5
Of course, you don’t want to make variable and function names unnecessary long (e.g., average would have been a great alternative function name, whereas computeAverageOfThisVector is probably too long). I like Ali’s principle:
Don’t minify your own code; use full variable names that the next developer can understand.
2. Write short functions that only do one thing
Ali argues “Functions are more understandable, readable, and maintainable if they do one thing only. If we have a bug when we write short functions, it is usually easier to find the source of that bug. Also, our code will be more reusable.” It thus helps to break up your code into custom functions that all do one thing and do that thing good!
For instance, our earlier function averageVector actually did two things. It first summated the vector, and then took the average. We can split this into two seperate functions in order to standardize our operations.
sumVector <- function(vector){
sum = 0
for(i in seq_along(vector)){
sum = sum + vector[i]
}
return(sum)
}
averageVector <- function(vector){
sum = sumVector(vector)
average = sum / length(vector)
return(average)
}
sumVector(1:10) # 55
averageVector(1:10) # 5.5
If you are writing a function that could be named with an “and” in it — it really should be two functions.
3. Documentation
Personally, I am terrible in commenting and documenting my work. I am always too much in a hurry, I tell myself. However, no more excuses! Anybody should make sure to write good documentation for their code so that future developers, including future you, understand what your code is doing and why!
Ali uses the following great example, of a piece of code with magic numbers in it.
areaOfCircle <- function(radius) {
return(3.14 * radius ** 2)
}
Now, you might immediately recognize the number Pi in this return statement, but others may not. And maybe you will need the value Pi somewhere else in your script as well, but you accidentally use three decimals the next time. Best to standardize and comment!
PI <- 3.14 # PI rounded to two decimal places
areaOfCircle <- function(radius) {
# Implements the mathematical equation for the area of a circle:
# Pi times the radius of the circle squared.
return(PI * radius ** 2)
}
The above is much clearer. And by making PI a variable, you make sure that you use the same value in other places in your script! Unfortunately, R doesn’t handle constants (unchangeable variables), but I try to denote my constants by using ALL CAPITAL variable names such as PI, MAX_GROUP_SIZE, or COLOR_EXPERIMENTAL_GROUP.
Do note that R has a built in variable pi for purposes such as the above.
I love Ali’s general rule that:
Your comments should describe the “why” of your code.
However, more elaborate R programming commenting guidelines are given in the Google R coding guide, stating that:
Functions should contain a comments section immediately below the function definition line. These comments should consist of a one-sentence description of the function; a list of the function’s arguments, denoted by
Args:, with a description of each (including the data type); and a description of the return value, denoted byReturns:. The comments should be descriptive enough that a caller can use the function without reading any of the function’s code.
Either way, prevent that your comments only denote “what” your code does:
# EXAMPLE OF BAD COMMENTING ####
PI <- 3.14 # PI
areaOfCircle <- function(radius) {
# custom function for area of circle
return(PI * radius ** 2) # radius squared times PI
}
5. Be Consistent
I do not have as strong a sentiment about consistency as Ali does in her article, but I do agree that it’s nice if code is at least somewhat in line with the common style guides. For R, I like to refer to my R resources list which includes several common style guides, such as Google’s or Hadley Wickham’s Advanced R style guide.












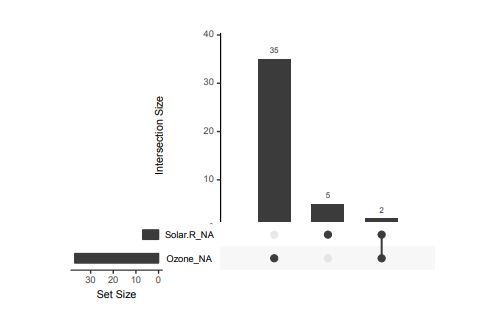
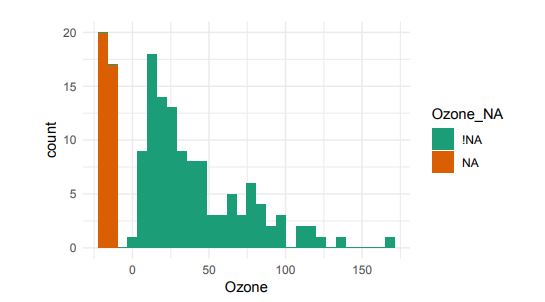 Alternatively, scatterplots can be easily generated. Displaying missings at 10 percent below the minimum of the airquality dataset. Scatterplots of ozone and solar radiation (A), and ozone and temperature (B). These plots demonstrate that there are missings in ozone and solar radiation, but not in temperature.
Alternatively, scatterplots can be easily generated. Displaying missings at 10 percent below the minimum of the airquality dataset. Scatterplots of ozone and solar radiation (A), and ozone and temperature (B). These plots demonstrate that there are missings in ozone and solar radiation, but not in temperature.